Automatic Data Augmentation Learning using Bilevel Optimization for Histopathological Images
Résumé
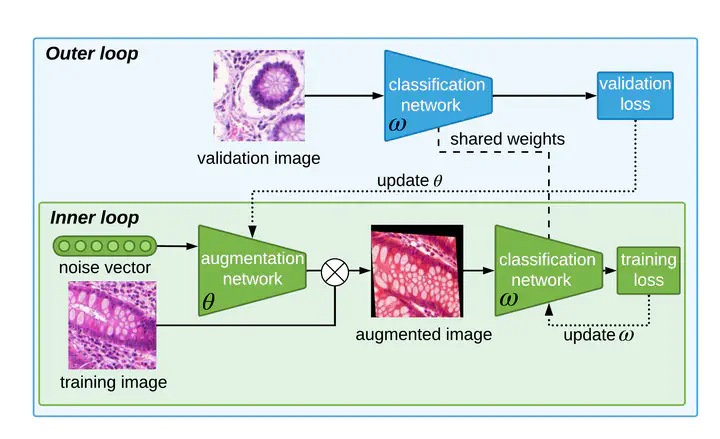
One of the main challenges faced when training a deep learning based model to classify histopathological images is the color and shape variability of the cells and tissues. Those variations can be due to the image acquisition process, more precisely the cell staining for color variations or tissue deformation for shape variations, or because the dataset is made of images from different devices. To tackle this challenge, Data Augmentation (DA) can be used during training to teach the model to be invariant to those color and shape variations. DA consists in creating new samples by applying transformations to existing ones. The problem with DA is that it is not only dataset specific but it also requires domain knowledge, which is not always available. Without this knowledge, selecting the right transformations can only be done using heuristics or with a computationally demanding search. To address this, we propose in this work an automatic DA learning method. In this method, the DA parameters, i.e. the transformation parameters needed to improve the model training, are considered learnable parameters and are learned automatically using a bilevel optimization approach in a quick and efficient way using truncated backpropagation. We validated the method on six different datasets of histopathological images. Experimental results show that our model can learn color and affine transformations that are more helpful to train an image classifier than predefined DA transformations. Predefined DA transformations are also more expensive as they need to be selected before the training by grid search on a validation set. We also show that similarly to a model trained with a RandAugment based framework, our model has also only a few method-specific hyperparameters to tune but is performing better. This makes our model a good solution to learn the best data augmentation parameters, especially in the context of histopathological images, where defining potentially useful transformation heuristically is not trivial.